Debugging Pandas Unexpected Type Conversion
Problem Statement:
While working with Pandas to process CSV files, I encountered a peculiar issue where certain columns, despite being explicitly defined as strings, were getting converted into float64, leading to unexpected NaN values after transformations. This post walks through our debugging process, the cause of the issue, and the solutions I found.
Explanation:
I was processing a dataset that included columns like ORDER_DATE, DELIVERY_DATE, and CUSTOMER_TYPE (these are just examples). The expectation was that all values in these columns would be treated as strings, because while creating the CSV I have given all the columns to be strings. However, when working with this CSV I checked the data types after reading the CSV, I observed an inconsistency:
Here’s the example dataset, which are strings:
get_csv = pd.read_csv(StringIO(csv_data))print(get_csv)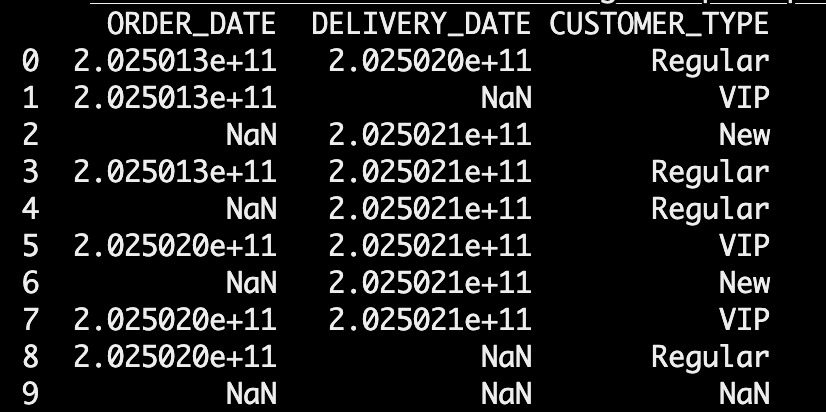
print(get_csv['CUSTOMER_TYPE'].apply(type).value_counts())print(get_csv['ORDER_DATE'].apply(type).value_counts())Output:
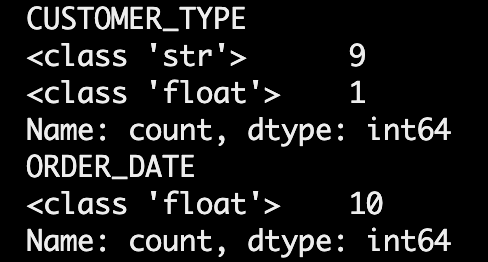
This showed that some values in ORDER_DATE and CUSTOMER_TYPE were being interpreted as floats, even though source CSV file has been explicitly set to strings.
Why Did This Happen?
Pandas automatically infer data types based on the values in the columns. Here’s why:
1. Pandas Converts Mixed Data Types Automatically
- If a column contains only strings and NaNs, Pandas should treat it as object (string).
However, if the column has numeric-looking strings + NaNs, Pandas assumes it’s a float column weird right!.
2. NaNs in a Mixed Column are Default to Float
When a column has mixed types (strings and NaN), Pandas coerces the entire column into float64 by default. (Completely new observation!!)
The Solution:
To prevent Pandas from making these incorrect assumptions, we have to explicitly enforced string types to the columns when reading the CSV:
1. Explicitly Set Data Types in read_csv()
get_csv = pd.read_csv( StringIO(csv_data), dtype={'ORDER_DATE': str, 'DELIVERY_DATE': str}, low_memory=False, escapechar='\\')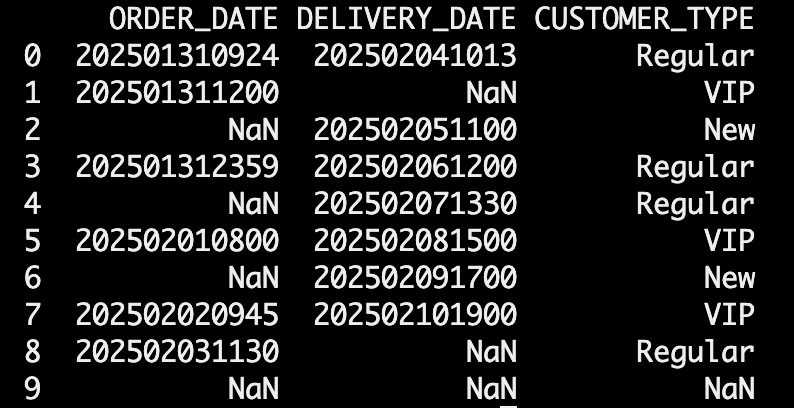
Or if you want to read that entire CSV should be read as string then we can directly give the parameter dtype to be string.
get_csv = pd.read_csv( StringIO(csv_data), dtype=str, low_memory=False, escapechar='\\')Now you there’s difference in the output!
✅ This ensures that Pandas does not infer the column types and treats them strictly as strings.
2. Convert to String After Loading
If the CSV was already read, we forced all values to be strings, ensuring that NaN values were handled correctly, we can use astype:
get_csv['ORDER_DATE'] = get_csv['ORDER_DATE'].astype(str)get_csv['CUSTOMER_TYPE'] = get_csv['CUSTOMER_TYPE'].astype(str)✅ This prevents Pandas from treating large numbers as floats.
Summary:
-
Pandas infers data types automatically, which can lead to unexpected conversions.
-
Large numeric-looking values mixed with NaNs may be interpreted as float64, leading to NaN issues.
-
Explicitly setting dtype in read_csv() prevents these issues.
-
Using .astype(str) and replacing unwanted NaNs ensures data consistency.
-
This debugging experience highlights the importance of carefully managing data types in Pandas, especially when dealing with CSV files containing mixed data formats. If you’re working with similar issues, try enforcing string types explicitly and always verify column types after loading your data!
Have you encountered similar Pandas quirks? Let me know how you handled them!
...
Further Reading
- The Lazy Solution! 02.May.2025
- Debugging Pandas Unexpected Type Conversion 07.Feb.2025
- Part-2 More about Docker 27.Feb.2023
...